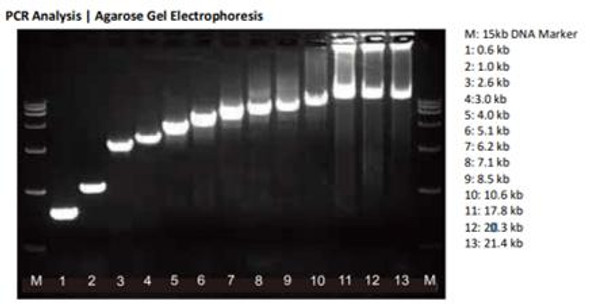
Description
Product Information
Genie Fusion Ultra High-Fidelity Red 2x Master Mix contains a next generation enzyme with a loading dye for robust PCR with ultra high-fidelity. With extremely low error rates x53-fold lower than Taq, Genie Fusion Ultra High-Fidelity DNA Polymerase sets a new level of performance for standard and long fragment PCR amplification.
Employing a unique extension factor as well as specificity-promoting and plateau-inhibiting factors, Genie Fusion greatly improves long range PCR amplification, specificity, and PCR yield. Genie Fusion is capable of amplifying long fragments such as 40 kb λ DNA, 40 kb plasmid DNA, 20 kb genomic DNA and 10 kb cDNA. In addition, Genie Fusion has excellent resistance to PCR inhibitors and can be used for direct PCR amplifications of bacteria, fungi, plant tissues, animal tissues, and even whole blood samples. Genie Fusion contains two monoclonal antibodies inhibiting the 5´→3´ polymerase activity and 3´→5´ exonuclease activity at room temperature which enables Genie Fusion to perform super-specific Hot-Start PCR.
Genie Fusion Ultra High-Fidelity Red 2X Master Mix contains Genie Fusion Ultra High-Fidelity DNA Polymerase, dNTPs and an optimized buffer system containing a loading dye. Protective agents in the Genie Fusion Ultra High-Fidelity Red 2X Master Mix allow for repeat freeze-thaw cycles while maintaining excellent performance. Amplification generates blunt-end products which are compatible with GenieClone DNA Assembly Cloning Kit (Assay Genie, Cat. No. #MORV0004).
Key Features
- Optimized 2x mastermix containing Ultra high-fidelity DNA polymerase, dNTPs and next-generation buffer system containing a loading dye
- Ultra high-fidelity DNA polymerase with error rates x53-fold lower than conventional Taq and x6-fold lower than Pfu
- Excels at amplifying long fragments such as 40 kb λ DNA, 40 kb plasmid DNA, 20 kb genomic DNA and 10 kb cDNA
- Superior performance with GC-rich templates
- Extreme resistance to PCR inhibitors
- Excellent choice for direct PCR from bacteria, fungi, plant tissues, animal tissues and whole blood samples
- Formulated with dual monoclonal antibodies for super specific Hotstart PCR
- Compatible with GenieClone DNA Assembly Cloning Kit (MORV0004)
2. Product Components
| Components | MORV0003 | MORV0003-5 | MORV0003-15 |
|
Genie Fusion Ultra High-Fidelity Red 2X Master Mix |
1 ml 40 x 50μl reactions |
5 x 1ml 200 x 50μl reactions |
15 x 1 ml 600 x 50μl reactions |
3. Storage
Store at -20℃; avoid repeated freezing and thawing.
4. Unit Definition
One unit (U) is defined as the amount of enzyme that incorporates 10 nmol of whole dNTPs into acid-insoluble products in 30 minutes at 74℃ with activated salmon sperm DNA as the template /primer.
5. Quality Control
Residual Endonulease Test: The product is tested in a reaction containing 25 μl of Genie Fusion Ultra High-Fidelity Red 2X Master Mix and 0.3 μg of Supercoiled pBR322 DNA. After incubation at 37℃ for 4 hours, there is no visually discernible change in DNA bands determined by agarose gel electrophoresis.
ResidualE.Coli gDNA Test: The residual nucleotide in 25 μl of Genie Fusion Ultra High-Fidelity Red 2X Master Mix is tested by SYBR® Red qPCR using specific primers of E.Coli gDNA. The residual E.Coli gDNA is lower than 10 copies.
Functional Assay 1: In a 50μl PCR system with 1U of Genie Fusion Ultra High-Fidelity Red 2X Master Mix, 100 ng of human genomic DNA was used as template. After 35 cycles, 1/10 of PCR products were detected by 1% agarose gel electrophoresis. A single DNA band of 8.2 kb was detected after EB staining.
Functional Assay 2: In a 50μl PCR system with 1U of Genie Fusion Ultra High-Fidelity Red 2X Master Mix, 10 ng of λ DNA was used as template. After 30 cycles, 1/10 of PCR products were detected by 1% agarose gel electrophoresis. A single DNA band of 15 kb was detected after EB staining.
6. Experimental Process
6.1 Standard PCR
Recommended PCR System
Keep all components on ice during the experiment. Mix components thoroughly after thawing and replace at -20℃ immediately use.
|
DdH2O |
up to 50 μl |
|
Genie Fusion Ultra High-Fidelity Red 2X Master Mix |
25 μl |
|
rimer 1 (10 μM) |
5 μl |
|
Primer 2 (10 μM) |
5 μl |
|
Template DNAª |
x μl |
The Assay Genie PCR Enhancer is recommended for unsuccessful amplification of fragments with GC content > 60%.
a. Optimal reaction concentration varies in different templates. In a 50 µl system, the recommended template usage is as follows:
|
Templates |
Input Template DNA |
|
Genomic DNA |
50 - 400 ng |
|
Plasmid or Virus DNA |
10 pg- 30 ng |
|
cDNA |
1 - 5 μl (≤ 1/10 of the total volume of PCR system) |
Recommended PCR Program
|
Steps |
Temperature |
Time |
Cycles |
|
Pre-denaturation a |
95℃ |
30 sec / 3 min |
1 |
|
Denaturation |
95℃ |
15 sec |
} 25 - 35 |
|
Annealing b |
56-72℃ |
15 sec |
|
|
Extension c |
72℃ |
30 - 60 sec / kb |
|
|
Final Extension |
72℃ |
5 min |
1 |
a. For pre-denaturation, the recommended temperature is 95℃, and the recommended time is 30 sec for plasmid / virus DNA and 3 min for genomic DNA / cDNA.
b. For annealing, the recommended temperature is the Tm of the primers. If the Tm of the primers is higher than 72℃, the annealing step can be removed (two-step PCR). If necessary, annealing temperature can be further optimized in a gradient. In addition, the amplification specificity depends directly on the annealing temperature. Raising annealing temperature is helpful to improve poor amplification specificity.
c. Longer extension time is helpful to increase the amplification yield.
6.2 For Long-fragment PCR
Genie Fusion Ultra High-Fidelity DNA Polymerase can perform a long-range amplification with high specificity and yields. If the standard PCR protocol (Section 6.1) yields insufficient results, the following Touch-Down, two-step PCR is recommended.
|
Steps |
Temperature |
Time |
Cycles |
|
Pre-denaturation |
95℃ |
3 min |
1 |
|
Denaturation |
92℃ |
15 sec |
} 5 |
|
Extension |
74℃ |
60 sec/kb |
|
|
Denaturation |
95℃ |
15 sec |
} 5 |
|
Extension |
72℃ |
60 sec/kb |
|
|
Denaturation |
95℃ |
15 sec |
} 5 |
|
Extension |
70℃ |
60 sec/kb |
|
|
Denaturation |
95℃ |
15 sec |
} 25 |
|
Extension |
68℃ |
60 sec/kb |
|
|
Final Extension |
68℃ |
5 min |
1 |
It is recommended to use high-quality templates and long primers . Increasing the input of template DNA may be helpful to improve the amplification yield.
6.3 Direct PCR with Crude Samples
Genie Fusion has excellent resistance to PCR inhibitors and can be used for direct PCR amplification of bacteria, fungi, plant tissues, animal tissues, and even whole blood samples. Crude materials that have been successfully amplified with Genie Fusion are as follows:
|
Sample Type |
Amplification Method |
Temperature Recommendation (for a 50 μl PCR system) |
|
Whole Blood |
Direct PCR |
1 - 5 µl |
|
Filter Paper Dry Blood |
Direct PCR |
1 - 2 mm2 filter paper |
|
Cultured Cells |
Direct PCR |
Low concentration of cells |
|
Yeast |
Direct PCR |
Single clone or 1 µl suspension |
|
Bacteria |
Direct PCR |
Single clone or 1 µl suspension |
|
Mod |
Direct PCR |
Low concentration of sample |
|
Sperm |
Direct PCR |
Low concentration of sample |
|
Plankton |
Direct PCR |
Low concentration of sample |
|
Plant Tissue |
Direct PCR |
1 - 2 mm2 tissue |
|
Mouse Tail |
PCR with lysate |
1 - 5 µl lysate* |
|
Food |
PCR with lysate |
1 - 5 µl lysate* |
*Lysate Preparation:
|
Animal Tissues or Food samples |
➜ |
Submerge little mount of samples in lysis buffer with final concentration 200 µg/ml of Proteinase K (self-provide). |
➜ |
60℃ 10 min 95℃ 10 min |
➜ |
Mix well and spin at room temperature. Collect the supernatant as lysate. |
Lysis Buffer: 20 mM of Tris-HCl, 100 mM of EDTA, 0.1% SDS, pH 8.0 (not included in this kit).
7. Application Examples
Note: See technical manual for application examples.
8. Notes
- Use high-quality templates.
- Do not use dUTP or any primers or templates that contain uracil.
- The Genie Fusion Ultra High-Fidelity Polymerase has strong proofreading activity. Therefore, the PCR products must be purified before adding A-Tailing when TA cloning.
- Primers design notes:
- Choose C or G as the last base of the 3’-end of the primer.
- Avoid continuous mismatching at the last 8 bases of the 3’-end of the primer.
- Avoid hairpin structures at the 3’-end of the primer.
- Tm of the primers should be within the range of 55℃ - 65℃ (recommend to calculate in Primer Premier 5), and the Tm difference between F and R primers should be less than 1℃.
- Additional sequence should not be included when calculating Tm of the primers.
- GC content of the primers should be within the range of 40% - 60%.
- The general distribution of A, G, T, C in the primers should be uniform, and avoid using regions with rich GC and rich AT.
- Keep complementary sequence less than 5 bases within the primers or between two primers, and complementary sequence less than 3 bases at the 3’-end of the primers.
- Please search the specificity of the designed primers by NCBI BLAST to avoid non-specific amplification.
9. Troubleshooting
No or Low Yield of PCR Products
|
Primers |
Optimize primer design |
|
Annealing Temperature |
Set gradient annealing temperature to find out the optimal one |
|
Concentration of Primers |
Optimize the concentration of primers |
|
Extension Time |
Optimize the extension time to 30 sec/kb-1 min/kb |
|
Cycle Numbers |
Increase cycle numbers to 35 - 40 |
|
Purity of Templates |
Use high-purity templates |
|
Template Input |
Refer to the recommended reaction system and increase the input |
Unspecific or Smear Bands in Electrophoresis
|
Primers |
Optimize primer design |
|
Annealing Temperature |
Try to improve annealing temperature and set gradient annealing temperature to optimize |
|
Concentration of Primers |
Decrease the concentration of primers to final concentration as 0.2 µM |
|
Extension Time |
Appropriately decrease the extension time when bands longer than target bands appear |
|
Cycle Numbers |
Decrease cycle number to 25 - 30 |
|
PCR Programs |
Use Two-Step PCR or Touch-down PCR |
|
Purity of Templates |
Use high purity templates |
|
Template Input |
Modify or decrease templates input referring to the recommended reaction system |