Comprehensive Analysis of Antibody Structure and Function
Antibodies, or immunoglobulins, stand as critical components of the immune system, orchestrating the identification and neutralization of pathogens like viruses and bacteria. This extensive article aims to provide a thorough understanding of the sophisticated architecture and multifaceted roles of antibodies, delving into their molecular composition, mechanisms behind their diversity and specificity, and their integral functions within the immune response.
Fundamental Architecture of Antibodies
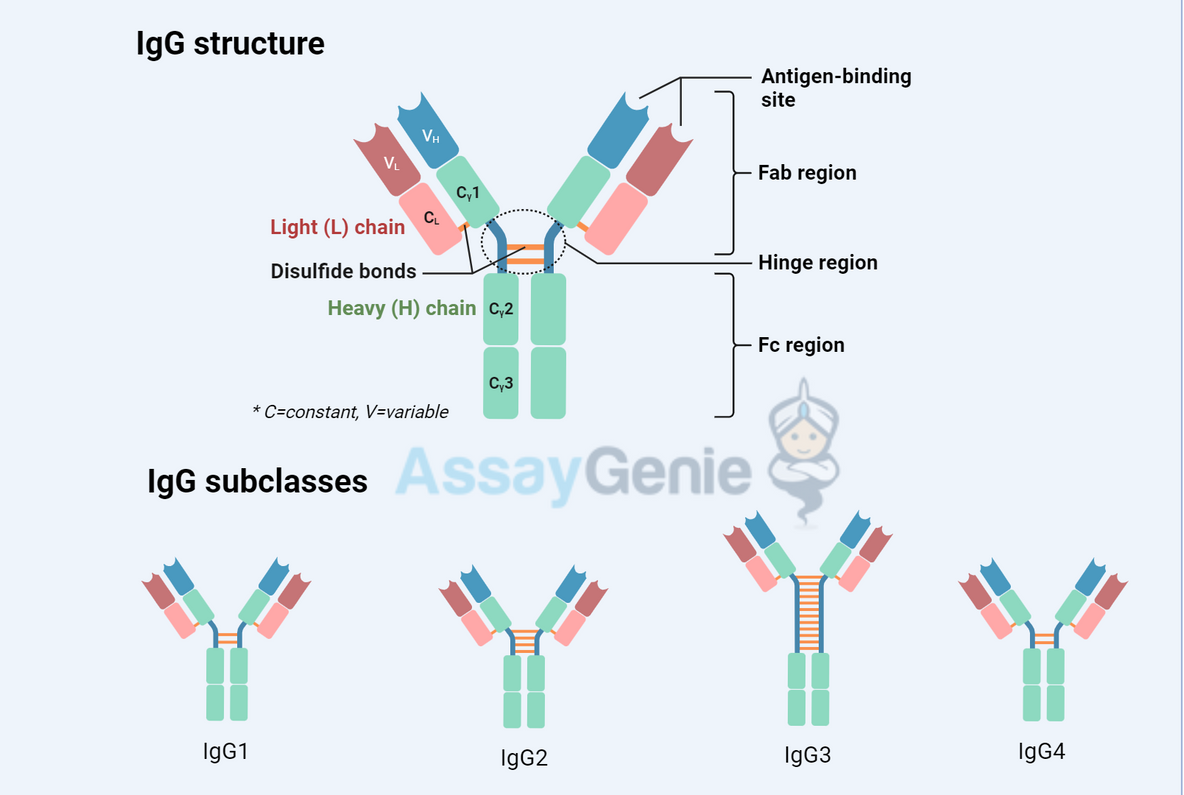
Antibodies are Y-shaped molecules composed of four polypeptide chains: two identical heavy chains and two identical light chains. These chains are linked together by disulfide bonds and non-covalent interactions, forming a structure that is both stable and flexible, allowing for efficient antigen recognition and binding.
Structural Components of Antibodies
Heavy and Light Chains: The Structural Backbone
Variable and Constant Regions: The Basis for Diversity and Function
- Variable (Fab) Region: Located at the tips of the Y-shaped structure, this region is responsible for antigen binding. The variability in the amino acid sequences within this region allows antibodies to recognize a vast array of antigens.
- Constant (Fc) Region: This region determines the antibody's isotype and mediates various effector functions by interacting with different components of the immune system.
Isotype Switching: Adapting the Immune Response
Isotype switching is a mechanism that allows an antibody to change its constant region, thereby altering its effector functions without affecting its antigen specificity. This process enables the immune system to tailor its response to different stages of infection and various types of pathogens.
Table 1: Antibody Isotypes and Their Functions
| Isotype | Function | Location |
| IgG | Systemic immunity, neutralization, opsonization | Blood, extracellular fluid |
| IgA | Mucosal immunity, neutralization | Mucosal areas |
| IgM | Early immune response, agglutination | B cell surface, blood |
| IgE | Allergy, defense against parasites | Bound to mast cells and basophils |
| IgD | B cell receptor | B cell surface |
Mechanisms Behind Antibody Diversity
The human immune system can produce an immense variety of antibodies, each with the potential to recognize a distinct antigen. This diversity is generated through several genetic and molecular mechanisms:
V(D)J Recombination and Somatic Hypermutation: The Engines of Diversity
V(D)J Recombination: The Genetic Shuffle
V(D)J recombination is the primary mechanism of diversity in the antibody variable regions, randomly assembling different V (Variable), D (Diversity), and J (Joining) gene segments. This recombination occurs in the heavy chain locus for both B and T cells, and in the light chain locus for B cells only, laying the foundation for antigen specificity.
Somatic Hypermutation and Affinity Maturation
Following initial antigen exposure, somatic hypermutation targets the variable regions of the antibody gene, introducing mutations that can increase or decrease affinity for the antigen. B cells with higher-affinity antibodies receive survival and proliferation signals, leading to affinity maturation over time.
Table 2: Mechanisms of Antibody Diversity
| Mechanism | Description |
| V(D)J Recombination | Random assembly of V, D, and J gene segments. |
| Somatic Hypermutation | Mutations in variable region genes increase affinity. |
| Isotype Switching | Changes in constant region modify effector functions. |
Complementarity Determining Regions (CDRs): Key to Antigen Recognition
The antigen-binding site of an antibody is formed by the variable domains of the heavy and light chains, with three complementarity-determining regions (CDRs) in each domain playing a critical role in antigen recognition. CDR3 is particularly significant due to its high variability, directly contributing to the specificity of antigen binding.
The Multifaceted Roles of Antibodies in Immune Response
Antibodies do much more than simply bind to antigens; they initiate a cascade of immune responses that help to neutralize and eliminate pathogens.
Effector Functions Mediated by the Constant Region
The Fc region of antibodies engages with Fc receptors on the surface of immune cells, triggering various effector functions:
Neutralization: Blocking pathogens from infecting host cells.
Opsonization: Marking pathogens for destruction by phagocytes.
Complement Activation: Initiating the complement cascade, leading to the lysis of pathogens.
Antibody-Dependent Cellular Cytotoxicity (ADCC): Recruiting natural killer (NK) cells to destroy antibody-coated targets.
Table 3: Antibody Effector Functions
| Function | Description |
| Neutralization | Blocking pathogens or toxins. |
| Opsonization | Facilitating phagocytosis. |
| Complement Activation | Triggering pathogen lysis. |
| ADCC | Destroying antibody-coated targets. |
Advanced Understanding of Antibody Function
Antibodies do not operate in isolation; their interactions with other components of the immune system amplify their effectiveness. The Fc region's engagement with Fc receptors on immune cells triggers a cascade of responses, including phagocytosis, antibody-dependent cellular cytotoxicity (ADCC), and complement activation, illustrating the antibody's role as a bridge between innate and adaptive immunity.
Conclusion
References
- Murphy, K., Travers, P., Walport, M., & Janeway, C. (2017). Janeway's Immunobiology (9th ed.). Garland Science. A foundational textbook providing in-depth coverage of immunological principles and mechanisms, including the structure and function of antibodies.
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2014). Molecular Biology of the Cell (6th ed.). Garland Science. Offers a comprehensive overview of cellular mechanisms, including the immune system's molecular basis.
- Tonegawa, S. (1983). Somatic generation of antibody diversity. Nature, 302(5909), 575-581.
- Rajewsky, K. (1996). Clonal selection and learning in the antibody system. Nature, 381(6585), 751-758.
- Nimmerjahn, F., & Ravetch, J. V. (2008). Fcγ receptors as regulators of immune responses. Nature Reviews Immunology, 8(1), 34-47.
- Schmitz, R., Young, R. M., Ceribelli, M., Jhavar, S., Xiao, W., Zhang, M., Wright, G., Shaffer, A. L., Hodson, D. J., Buras, E., Liu, X., Powell, J., Yang, Y., Xu, W., Zhao, H., Kohlhammer, H., Rosenwald, A., Kluin, P., Muller-Hermelink, H. K., Ott, G., Gascoyne, R. D., Connors, J. M., Rimsza, L. M., Campo, E., Jaffe, E. S., Delabie, J., Smeland, E. B., Ogwang, M. D., Reynolds, S. J., Fisher, R. I., Braziel, R. M., Kridel, R., Chan, W. C., Weisenburger, D. D., & Staudt, L. M. (2012). Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature, 490(7418), 116-120.
- Abbas, A. K., Lichtman, A. H., & Pillai, S. (2020). Cellular and Molecular Immunology (10th ed.). Elsevier.
- Hozumi, N., & Tonegawa, S. (1976). Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proceedings of the National Academy of Sciences, 73(10), 3628-3632.
Written by Zainab Riaz
Zainab Riaz completed her Master degree in Zoology from Fatimah Jinnah University in Pakistan and is currently pursuing a Doctor of Philosophy in Zoology at University of Lahore in Pakistan.
Recent Posts
-
Growth Factors Can Cooperate to Promote Tumorigenesis
Introduction Tumorigenesis, the process by which normal cells transform into cancer cells …3rd May 2024 -
Inflammasome Activation Pathways: A Comprehensive Overview
Inflammasomes are complex intracellular structures that play a pivotal role in the immune respon …29th Apr 2024 -
Illuminating the Multifaceted Role of Acetylation: Bridging Chemistry and Biology Introduction:
Acetylation, a chemical process characterized by the addition of an acetyl functional group t …16th Apr 2024