Targeted Protein Degradation: The Next Frontier in Oncology Drug Discovery
For decades, cancer drug development has focused on blocking proteins—inhibiting enzymes, antagonizing receptors, disrupting signaling pathways. Yet this approach leaves a vast landscape of disease-driving proteins untouched, particularly those without enzymatic activity or binding pockets suitable for traditional small molecules. Now, a fundamentally different strategy is reshaping oncology: instead of merely inhibiting proteins, we can eliminate them entirely through targeted protein degradation.
Introduction
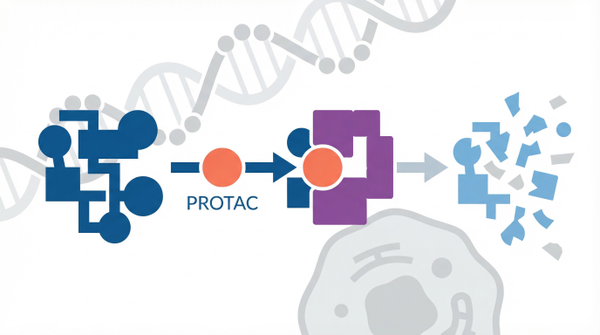
The concept of targeted protein degradation (TPD) represents a paradigm shift in how we approach cancer therapy. Rather than occupying a protein's active site to block its function, TPD technologies hijack the cell's own protein disposal machinery—the ubiquitin-proteasome system—to selectively destroy disease-causing proteins. This approach offers several compelling advantages over conventional inhibitors: it can target proteins previously considered "undruggable," it operates catalytically rather than occupancy-driven (meaning lower doses may achieve therapeutic effects), and it can overcome resistance mechanisms that plague traditional inhibitors. The field has evolved rapidly from academic curiosity to clinical reality, with emerging therapeutic modalities tackling undruggable proteins now advancing through clinical trials. As we stand at this inflection point, understanding the mechanisms, opportunities, and challenges of protein degradation is essential for anyone involved in cancer drug discovery.
From Molecular Glue to Designer Degraders: The Evolution of TPD
The journey of targeted protein degradation began serendipitously with the discovery that thalidomide and its derivatives—drugs initially developed for other purposes—could induce the degradation of specific transcription factors by recruiting them to the E3 ubiquitin ligase cereblon. This "molecular glue" mechanism revealed that small molecules could reprogram the substrate specificity of the cellular degradation machinery. Building on this insight, researchers developed proteolysis-targeting chimeras (PROTACs), bifunctional molecules that physically link a target protein to an E3 ligase, forming a ternary complex that tags the target for destruction. However, the field has expanded far beyond these initial modalities. Recent work has demonstrated that induced polymerization facilitates specific protein degradation, revealing an entirely distinct mechanism where small molecules trigger the reversible aggregation of target proteins, leading to their sequestration and subsequent degradation. This mechanistic diversity suggests that protein degradation is not a single technology but rather a versatile platform with multiple strategies for eliminating disease-causing proteins.
Conquering KRAS: A Watershed Moment for Degrader Technology
Perhaps no target better illustrates the transformative potential of protein degradation than KRAS, the most frequently mutated oncogene in human cancer. For decades, KRAS was considered the "holy grail" of undruggable targets—a small GTPase lacking obvious binding pockets and cycling rapidly between active and inactive states. While covalent inhibitors targeting the G12C mutation have achieved clinical success, they address only a fraction of KRAS-driven cancers. The development of small-molecule degraders addressing oncogenic KRAS alleles represents a watershed moment for the field. These pan-KRAS degraders can eliminate 13 of the 17 most common oncogenic KRAS variants, demonstrating that degradation can succeed where inhibition fails. Importantly, degrading KRAS produces more profound and sustained pathway suppression than inhibiting it, killing cancer cells while sparing normal tissues. The pharmacological degradation was well-tolerated in preclinical models and led to tumor regression, validating the therapeutic potential of this approach. This breakthrough suggests that many proteins dismissed as undruggable may become tractable through degradation strategies.
Expanding the Degrader Toolbox: From Small Molecules to Antibodies
While small-molecule degraders have dominated early development, the field is rapidly diversifying its technological arsenal. One critical limitation of traditional PROTACs is their inability to target cell-surface receptors, which lack access to intracellular E3 ligases. Researchers have overcome this barrier through the development of proteolysis-targeting antibodies (PROTABs), which enable tissue-selective cell-surface protein degradation by tethering transmembrane proteins to cell-surface E3 ubiquitin ligases like ZNRF3. This approach enables colorectal cancer-specific degradation and demonstrates that the degrader concept can be adapted to virtually any subcellular compartment. Meanwhile, systematic efforts to map the "degradable kinome" have provided chemical leads for approximately 200 kinases, with chemo-proteomics datasets fueling kinase degrader discovery and revealing that kinase degradation is p97-dependent. These expansive datasets demonstrate that starting from the highest-affinity binder is often an ineffective strategy for degrader development, highlighting the need for empirical screening and mechanistic understanding rather than simple extrapolation from inhibitor programs.
Precision Medicine Through Degradation: Context-Dependent Vulnerabilities
One of the most exciting aspects of protein degradation is its potential to exploit context-dependent vulnerabilities in cancer. Because degraders can eliminate proteins entirely rather than merely inhibiting them, they can reveal synthetic lethal relationships and redundancies that are invisible to traditional pharmacology. For instance, studies using chemical genetic systems to achieve acute protein shutdown have shown that CDC7 kinase has redundant roles with CDK1 during cell cycle progression, fundamentally revising our understanding of cell division control. More broadly, paediatric cancer vulnerabilities inform degrader design by identifying transcription factors and chromatin regulators that are essential in specific developmental contexts. This precision extends to tissue selectivity as well—by choosing E3 ligases with restricted expression patterns or exploiting tumor-specific protein-protein interactions, degraders can achieve selectivity that is difficult or impossible with traditional inhibitors. The ability to eliminate proteins in a tissue-specific, temporally controlled manner opens new therapeutic windows that were previously inaccessible.
From Bench to Bedside: Clinical Progress and Future Directions
The clinical translation of protein degraders has accelerated dramatically in recent years. Multiple PROTAC molecules have entered clinical trials for hematologic malignancies and solid tumors, with early results suggesting that the unique pharmacology of degraders translates into clinical benefit. The comprehensive review of recent progress in degrader modalities highlights that PROTACs are on the verge of their first clinical approval, with lenalidomide and related molecular glues already demonstrating the therapeutic potential of induced protein degradation. However, significant challenges remain. The molecular weight and physicochemical properties of bifunctional degraders can limit oral bioavailability and tissue penetration. The vast majority of the approximately 600 human E3 ligases remain unexploited, representing enormous untapped potential but also requiring substantial investment in ligand discovery. Resistance mechanisms are beginning to emerge, including mutations in E3 ligases or alterations in the ubiquitin-proteasome system itself. Addressing these challenges will require continued innovation in degrader design, deeper mechanistic understanding of the ubiquitination machinery, and careful attention to pharmacokinetic optimization.
A New Chapter in Cancer Therapeutics
Targeted protein degradation represents more than an incremental advance in cancer drug discovery—it is a fundamental reconceptualization of how small molecules can modulate biology. By eliminating disease-causing proteins rather than merely inhibiting them, degraders access a therapeutic space that was previously unreachable. The rapid progression from molecular glue discoveries to rationally designed pan-KRAS degraders demonstrates the field's maturation, while the expansion into antibody-based formats and novel mechanisms like induced polymerization reveals its continued innovation. As degraders move through clinical trials and the first approvals approach, we are witnessing the emergence of a new pharmacological modality that will likely transform oncology and extend far beyond cancer into other therapeutic areas. The proteins we once considered undruggable are now becoming druggable through degradation, opening a vast new frontier in precision medicine.
References
- Hinterndorfer M, Spiteri VA, Ciulli A, Winter GE (2025). Targeted protein degradation for cancer therapy. Nat Rev Cancer. 25(7):493-516. PMID: 40281114
- Popow J, Farnaby W, Gollner A, Kofink C, et al. (2024). Targeting cancer with small-molecule pan-KRAS degraders. Science. 385(6715):1338-1347. PMID: 39298590
- Békés M, Langley DR, Crews CM (2022). PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 21(3):181-200. PMID: 35042991
- Słabicki M, Yoon H, Koeppel J, Nitsch L, et al. (2020). Small-molecule-induced polymerization triggers degradation of BCL6. Nature. 588(7836):164-168. PMID: 33208943
- Donovan KA, Ferguson FM, Bushman JW, Eleuteri NA, et al. (2020). Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell. 183(6):1714-1731.e10. PMID: 33275901
- Marei H, Tsai WK, Kee YS, Ruiz K, et al. (2022). Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature. 610(7930):182-189. PMID: 36131013
- Chen X, Yang W, Roberts CWM, Zhang J (2024). Developmental origins shape the paediatric cancer genome. Nat Rev Cancer. 24(6):382-398. PMID: 38698126
- Suski JM, Ratnayeke N, Braun M, Zhang T, et al. (2022). CDC7-independent G1/S transition revealed by targeted protein degradation. Nature. 605(7909):357-365. PMID: 35508654
Recent Posts
-
Targeted Protein Degradation: The Next Frontier in Oncology Drug Discovery
For decades, cancer drug development has focused on blocking proteins—inhibiting enzymes, antagoniz …18th Feb 2026 -
Neuroinflammation and Compulsive Behavior: The Role of Astrocytes in OCD
Imagine a mind trapped in a loop, endlessly repeating actions or thoughts, driven by an invisible f …18th Feb 2026 -
Metabolic Exhaustion: How Mitochondrial Dysfunction Sabotages CAR-T Cell Therapy in Solid Tumors
Imagine engineering a patient's own immune cells into precision-guided missiles against cancer—cells …8th Dec 2025