RNA Extraction for COVID-19 Testing
Testing for COVID-19
Since the start of the COVID-19 pandemic, the World Health Organisation have emphasised the crucial importance of viral testing. This has therefore led to a huge demand for high-throughput testing of throat and nasopharyngeal samples (Source). The greater number of patients being qualitatively tested for coronavirus, the more treatment can be provided, and the greater the reduction in the rapid spread of this virus.
Typically, reverse-transcriptase polymerase chain reaction (RT-PCR) is used for viral diagnostic testing and requires a nasopharyngeal swab for the viral sample. Nasopharyngeal swabs are required to obtain samples of respiratory secretions. They are collected by inserting a small swab with a stop tip into the nostril. This is then guided to the back of the nose, and will be twirled a few times to collect secretions. The sample can be then sent for testing.
RT-PCR is the process of converting RNA into cDNA. It uses RNA as a template instead of DNA, which is seen in the standard PCR method. First, the enzyme reverse transcriptase uses the RNA template to produce a complementary single-stranded DNA strand called cDNA. This process is known as reverse transcription (Source). After, the enzyme DNA polymerase is used to convert the single-stranded DNA molecule into double-stranded DNA which can be used as templates for a PCR reaction. This allows the double-stranded DNA to be copied and amplified exponentially with the use of primers and thermocycling . The value of RT-PCR is that it can be used to determine if an RNA species, such as SARS-CoV-2 is present in a sample.
RNA Extraction Principle
In order to execute RT-PCR, RNA extraction must be carried out first. RNA extraction is the purification of RNA from biological samples. RNA extraction is a delicate process, as cells and the environment secrete high concentrations of enzymes that destroy nucleic acids, therefore, the process must be carried out in a careful and quick manner.
Most kits contain sterile buffers, solutions that lyse cells in order to access the RNA within the cells and a way of separating the RNA from DNA, proteins and other macronutrients found in the sample of cells.
The RNA extraction kits in general have few steps. First, the patient’s sample is mixed with a solution that lyses the cells in order to release the genetic material. After, inactivation of RNAse activity, denaturation of nucleoprotein complexes and removal of contaminating DNA and proteins must occur in order to purify the RNA. The resulting cellular debris and the extracted RNA can then proceed to the RT-PCR step.
Top ELISA Kits
RNA Extraction Technique
The most common method for RNA extraction is listed below:
- Lyse and homogenise cells using centrifugation and buffers in order to access the RNA within the cells.
- Cells are then treated with a buffer to inactivate RNases that may interfere with the RNA extraction step.
- RNA within the sample is isolated and separated from other components within the cell via different methods. These include guanidinium thiocyanate-phenol-chloroform extraction, paramagnetic particle technology or silica-membrane based technology.
- The sample is centrifuged which separates the RNA from the other molecules within the cell and forms a pellet at the bottom of the vial.
- The pellet is stored in RNase-free water and the other materials of the cells are discarded. The RNA can then be used in RT-PCR if it passes certain criteria. These include, that the RNA should be free from protein, genomic DNA or enzyme inhibitors such as DNase. The RNA must also not be coated with any alcohol, such as ethanol, which may compromise the RT-PCR reaction (Source).
The three methods used for RNA extraction are Phenol-Guanidine Isothiocyanate (GITC)-based solutions, paramagnetic particle technology or silica-membrane based technology. The principle and method behind each of these methods are discussed below.
Organic extraction includes separating RNA from DNA by coating the sample in acidic solutions containing guanidinium thiocyanate, sodium acetate, phenol and chloroform. This is then followed by centrifugation, which physically separates the RNA from DNA and other proteins. The RNA remains in the upper aqueous phase, whilst the DNA and other proteins stay in the middle or settle to the bottom. Total RNA can then be recovered by transferring the upper aqueous phase containing the RNA into a new tube and precipitating the RNA by using isopropanol, a less concentrated version of ethanol. This sample is then centrifuged, and the RNA will form a pellet at the bottom of the tube and can be isolated.
In paramagnetic particle technology, the use of magnetic beads that have an individual coating that allows them to have affinity for specific molecules within a sample, such as DNA, RNA or proteins administrates the isolation and separation of different particles within a sample. The RNA only binds to the magnetic beads that have been appropriately coated, leaving any remaining contaminants in solution.
Silica-membrane based technology is a solid phase extraction method used to purify nucleic acids. RNA will bind to the silica under certain conditions, whilst other molecules such as proteins and DNA will not. After the cells have been lysed and then genetic material is accessible, a buffer solution and ethanol or isopropanol are added to the sample. This forms the binding solution. This solution is then centrifuged in a spin column. This forces the binding solution through a silica gel membrane that is in the spin column. If the conditions such as pH and salt concentrations of the binding solution are optimal, the RNA will bind to the silica gel as the solution passes through.
RNA Extraction used for COVID-19 Testing
SARS-CoV-2, the viral agent that causes the disease, COVID-19, is a positive-sense single stranded RNA virus. Therefore, in order to detect specific RNA molecules, hence specific RNA viruses such as SARS-CoV-2, the RNA molecules must first be converted to DNA molecules for PCR detection.
The most common method used for extraction of RNA of in an infected person's nasopharyngeal sample is organic extraction. As mentioned before, this method involves isolating the RNA using guanidinium thiocyanate-phenol-chloroform extraction. The chemicals used in this extraction degrade any proteins or cellular components by breaking down the hydrogen bonds within the molecules, whilst protecting DNA and RNA. The RNA is then separated from DNA with an acidic solution consisting of guanidinium thiocyanate, sodium acetate, phenol, and chloroform. After centrifugation, under acidic conditions, the RNA will remain in the upper aqueous phase of the solution, whilst the DNA and proteins remain in the middle and lower organic phase. The total RNA is then precipitated with isopropanol to remove any remaining salts or chemicals on the RNA (Source).
The RNA can then be stored in RNase-free water until used in RT-PCR.
Recent Posts
-
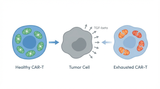
Metabolic Exhaustion: How Mitochondrial Dysfunction Sabotages CAR-T Cell Therapy in Solid Tumors
Imagine engineering a patient's own immune cells into precision-guided missiles against cancer—cells …8th Dec 2025 -
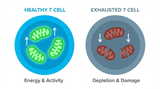
The Powerhouse of Immunity: How Mitochondrial Fitness Fuels the Fight Against Cancer
Why do powerful cancer immunotherapies work wonders for some patients but fail for others? The answe …5th Dec 2025 -
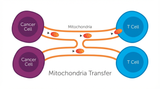
How Cancer Cells Hijack Immune Defenses Through Mitochondrial Transfer
Imagine a battlefield where the enemy doesn't just hide from soldiers—it actively sabotages their we …5th Dec 2025