Anti-SARS-CoV-2 Spike RBD Antibody (CAB20135)
- SKU:
- CAB20135
- Product Type:
- Antibody
- Antibody Type:
- Polyclonal Antibody
- Reactivity:
- Virus
- Host Species:
- Rabbit
- Isotype:
- IgG
Frequently bought together:
Description
| Product Name: | SARS-CoV-2 Spike RBD Rabbit pAb |
| Product Code: | CAB20135 |
| Size: | 20uL, 50uL, 100uL |
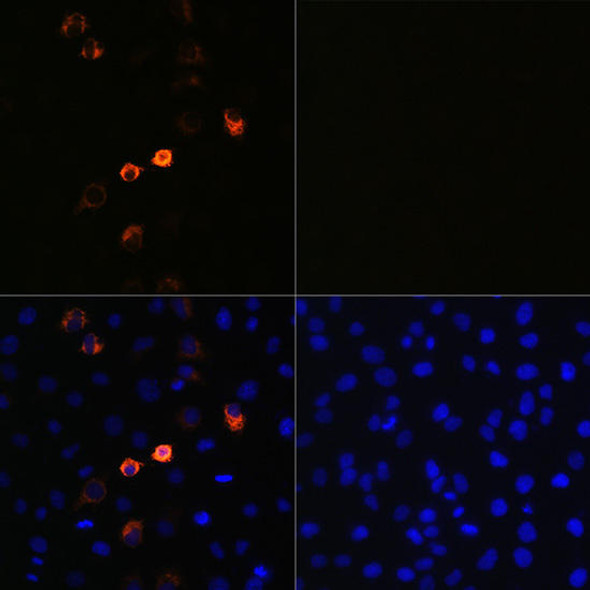
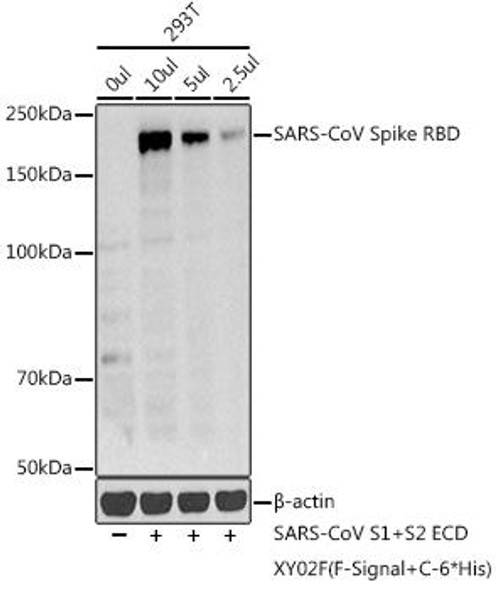
| Applications: | WB, IF, IP, ELISA |
| Reactivity: | SARS-CoV-2 |
| Host Species: | Rabbit |
| Immunogen: | Recombinant fusion protein of SARS-CoV-2 Spike RBD. |
| Applications: | WB, IF, IP, ELISA |
| Recommended Dilutions: | ELISA 1:50000-1:200000 WB 1:500 - 1:2000 IF 1:50 - 1:200 IP 1:50 - 1:200 |
| Reactivity: | SARS-CoV-2 |
| Positive Samples: | 293T |
| Immunogen: | Recombinant fusion protein of SARS-CoV-2 Spike RBD. |
| Purification Method: | Affinity purification |
| Storage: | Store at -20°C. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide, 50% glycerol, pH7.3. |
| Isotype: | IgG |
| Sequence: | Email for sequence |
| Gene ID: | 43740568 |
| Uniprot: | P0DTC2 |
| Observed MW: | 30kDa |
| UniProt Protein Function: | |
| UniProt Protein Details: | |
| NCBI Summary: | |
| UniProt Code: | P0DTC2 |
| NCBI GenInfo Identifier: | |
| NCBI Gene ID: | |
| NCBI Accession: | |
| UniProt Secondary Accession: | P0DTC2 |
| UniProt Related Accession: | P0DTC2 |
| Molecular Weight: | |
| NCBI Full Name: | |
| NCBI Synonym Full Names: | |
| NCBI Official Symbol: | |
| NCBI Official Synonym Symbols: | |
| NCBI Protein Information: | |
| UniProt Protein Name: | |
| UniProt Synonym Protein Names: | |
| Protein Family: | SMY2 |
| UniProt Gene Name: | |
| UniProt Entry Name: |


![Anti-SARS-CoV-2 Spike RBD [2165] Anti-SARS-CoV-2 Spike RBD [2165]](https://cdn11.bigcommerce.com/s-h68l9z2lnx/images/stencil/590x590/products/214648/569746/anti-sars-cov-2-spike-rbd-2165__70881.1673669701.jpg?c=2)
![Anti-SARS-CoV-2 Spike RBD [2838] Anti-SARS-CoV-2 Spike RBD [2838]](https://cdn11.bigcommerce.com/s-h68l9z2lnx/images/stencil/590x590/products/214651/569714/anti-sars-cov-2-spike-rbd-2838__09675.1673669671.jpg?c=2)
![Anti-SARS-CoV-2 Spike RBD [2355] Anti-SARS-CoV-2 Spike RBD [2355]](https://cdn11.bigcommerce.com/s-h68l9z2lnx/images/stencil/590x590/products/214681/569737/anti-sars-cov-2-spike-rbd-2355__83998.1673669692.jpg?c=2)